Kara’s first lead-author paper is out now in Molecular Ecology! Huge congrats to Kara and coauthors!
Link to the open-access study in Molecular Ecology.
Abstract
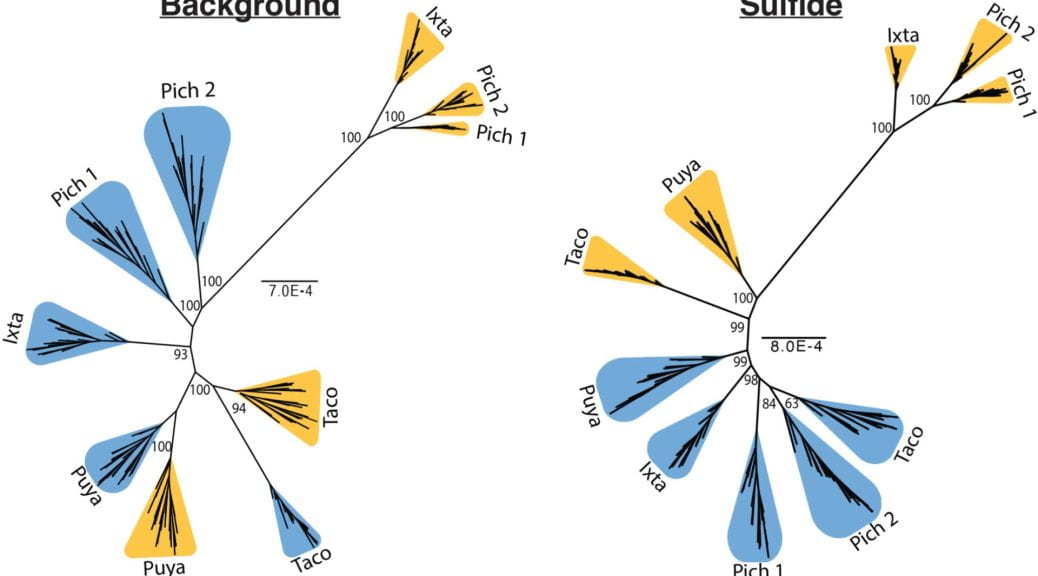
Hydrogen sulfide is a toxic gas that disrupts numerous biological processes, including energy production in the mitochondria, yet fish in the Poecilia mexicana species complex have independently evolved sulfide tolerance several times. Despite clear evidence for convergence at the phenotypic level in these fishes, it is unclear if the repeated evolution of hydrogen sulfide tolerance is the result of similar genomic changes. To address this gap, we used a targeted capture approach to sequence genes associated with sulfide processes and toxicity from five sulfidic and five nonsulfidic populations in the species complex. By comparing sequence variation in candidate genes to a reference set, we identified similar population structure and differentiation, suggesting that patterns of variation in most genes associated with sulfide processes and toxicity are due to demographic history and not selection. But the presence of tree discordance for a subset of genes suggests that several loci are evolving divergently between ecotypes. We identified two differentiation outlier genes that are associated with sulfide detoxification in the mitochondria that have signatures of selection in all five sulfidic populations. Further investigation into these regions identified long, shared haplotypes among sulfidic populations. Together, these results reveal that selection on standing genetic variation in putatively adaptive genes may be driving phenotypic convergence in this species complex.